class: center, middle, inverse, title-slide # tidy data and tidy tools ## Overview of tools of effective data analysis ### Eric Leung ### 2020-05-17 --- # Overview - Tidy data - Core and useful tidyverse functions - Useful packages --- # Tidy data overview  *Source*: https://r4ds.had.co.nz/tidy-data.html --- # Tidy data are easier to work with inside the tidyverse > "Tidy datasets are all alike, but every messy dataset is messy in its own way." > > –– Hadley Wickham --- # Objective Take built-in phyloseq object and convert it to a tidy data set. Then wrangle the data using tidy principles and tidyverse packages<sup>*</sup>. .footnote[[*] The "tidyverse" packages are a suite of packages that adhere to tidy data principles] --- # Load packages ```r library(phyloseq) # install.packages("remotes") # remotes::install_github("mikemc/speedyseq") library(speedyseq) # Optional ``` --- # phyloseq data example ```r # Load data example data("GlobalPatterns") GlobalPatterns ``` ``` ## phyloseq-class experiment-level object ## otu_table() OTU Table: [ 19216 taxa and 26 samples ]: ## sample_data() Sample Data: [ 26 samples by 7 sample variables ]: ## tax_table() Taxonomy Table: [ 19216 taxa by 7 taxonomic ranks ]: ## phy_tree() Phylogenetic Tree: [ 19216 tips and 19215 internal nodes ]: ## taxa are rows ``` -- Let's convert this into a tidy data set. --- # `psmelt()` converts to tidy data ```r psm_gp <- psmelt(GlobalPatterns) # Create tidy data ``` -- ```r library(tibble) psm_gp <- tibble(psm_gp) # Make nicer data frame ``` -- ```r head(psm_gp, 5) # View first five rows ``` ``` ## # A tibble: 5 x 17 ## OTU Sample Abundance X.SampleID Primer Final_Barcode Barcode_truncated_p… ## <chr> <chr> <dbl> <fct> <fct> <fct> <fct> ## 1 549656 AQC4cm 1177685 AQC4cm ILBC_… ACAGCT AGCTGT ## 2 279599 LMEpi24M 914209 LMEpi24M ILBC_… ACACTG CAGTGT ## 3 549656 AQC7cm 711043 AQC7cm ILBC_… ACAGTG CACTGT ## 4 549656 AQC1cm 554198 AQC1cm ILBC_… ACAGCA TGCTGT ## 5 360229 M31Tong 540850 M31Tong ILBC_… ACACGA TCGTGT ## # … with 10 more variables: Barcode_full_length <fct>, SampleType <fct>, ## # Description <fct>, Kingdom <chr>, Phylum <chr>, Class <chr>, Order <chr>, ## # Family <chr>, Genus <chr>, Species <chr> ``` --- # Quick note on the tidyverse - The {tidyverse} package is a suite of packages - Home website https://www.tidyverse.org/ - Running `library(tidyverse)` will load the main tidyverse packages - The following code loads the appropriate packages within the tidyverse when needed --- # Let's clean the `Descriptions` column ```r library(dplyr) psm_gp %>% select(OTU, Sample, SampleType, Description) %>% head(5) ``` ``` ## # A tibble: 5 x 4 ## OTU Sample SampleType Description ## <chr> <chr> <fct> <fct> ## 1 549656 AQC4cm Freshwater (creek) "Allequash Creek, 3-4 cm depth" ## 2 279599 LMEpi24M Freshwater "Lake Mendota Minnesota, 24 meter epilimni… ## 3 549656 AQC7cm Freshwater (creek) "Allequash Creek, 6-7 cm depth" ## 4 549656 AQC1cm Freshwater (creek) "Allequash Creek, 0-1cm depth" ## 5 360229 M31Tong Tongue "M3, Day 1, tongue, whole body study " ``` --- # Just keep stool samples ```r psm_gp %>% select(OTU, Sample, SampleType, Description) %>% filter(SampleType == "Feces") %>% head(5) ``` ``` ## # A tibble: 5 x 4 ## OTU Sample SampleType Description ## <chr> <chr> <fct> <fct> ## 1 331820 M11Fcsw Feces "M1, Day 1, fecal swab, whole body study " ## 2 331820 M31Fcsw Feces "M3, Day 1, fecal swab, whole body study" ## 3 189047 TS29 Feces "Twin #2" ## 4 158660 M11Fcsw Feces "M1, Day 1, fecal swab, whole body study " ## 5 244304 M11Fcsw Feces "M1, Day 1, fecal swab, whole body study " ``` -- The descriptions have lots of information in them that we might want to visualize later on with ggplot2. -- Difficult to plot when the data is locked in text form. **Note**: some of the following code may not be "best" design decision --- # Load core data wrangling libraries ```r library(tidyr) ``` -- ```r # Column must be character, `Description` is currently a factor psm_gp %>% select(OTU, Sample, SampleType, Description) %>% filter(SampleType == "Feces") %>% * mutate(Description = as.character(Description)) %>% * separate_rows(Description, sep = ", ") %>% head(5) ``` ``` ## # A tibble: 5 x 4 ## OTU Sample SampleType Description ## <chr> <chr> <fct> <chr> ## 1 331820 M11Fcsw Feces "M1" ## 2 331820 M11Fcsw Feces "Day 1" ## 3 331820 M11Fcsw Feces "fecal swab" ## 4 331820 M11Fcsw Feces "whole body study " ## 5 331820 M31Fcsw Feces "M3" ``` --- # Let's move values into columns ```r # Create `value` column to keep value psm_gp %>% select(OTU, Sample, SampleType, Description) %>% filter(SampleType == "Feces") %>% mutate(Description = as.character(Description)) %>% separate_rows(Description, sep = ", ") %>% * mutate(value = 1) %>% * pivot_wider(names_from = Description, * values_from = value) %>% head(5) ``` ``` ## # A tibble: 5 x 11 ## OTU Sample SampleType M1 `Day 1` `fecal swab` `whole body study ` M3 ## <chr> <chr> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> ## 1 331820 M11Fcsw Feces 1 1 1 1 NA ## 2 331820 M31Fcsw Feces NA 1 1 NA 1 ## 3 189047 TS29 Feces NA NA NA NA NA ## 4 158660 M11Fcsw Feces 1 1 1 1 NA ## 5 244304 M11Fcsw Feces 1 1 1 1 NA ## # … with 3 more variables: whole body study <dbl>, Twin #2 <dbl>, Twin #1 <dbl> ``` --- # We can reverse our pivot ```r psm_gp %>% select(OTU, Sample, SampleType, Description) %>% filter(SampleType == "Feces") %>% mutate(Description = as.character(Description)) %>% separate_rows(Description, sep = ", ") %>% mutate(value = 1) %>% pivot_wider(names_from = Description, values_from = value) %>% * pivot_longer(cols = !c(OTU, Sample, SampleType), * names_to = "Description", * values_to = "Value") %>% head(5) ``` ``` ## # A tibble: 5 x 5 ## OTU Sample SampleType Description Value ## <chr> <chr> <fct> <chr> <dbl> ## 1 331820 M11Fcsw Feces "M1" 1 ## 2 331820 M11Fcsw Feces "Day 1" 1 ## 3 331820 M11Fcsw Feces "fecal swab" 1 ## 4 331820 M11Fcsw Feces "whole body study " 1 ## 5 331820 M11Fcsw Feces "M3" NA ``` --- # Future steps - With the code here, you could further process the data to filter on specific features. - For example, you could focus on stool samples from a specific twins or a whole body study. - The {stringr} package within the tidyverse will prove useful for checking the contents of strings --- # Functions reviewed - **Note**: the following functions are in the form `<package>::<function>` - **`dplyr::select()`** = select, subset, and remove columns from data frame - **`dplyr::filter()`** = remove or keep specific rows based on information in data frame - **`dplyr::mutate()`** = convert or transform column values - **`tidyr::separate_rows()`** = take column values and separate them into multiple rows - **`tidyr:pivot_wider()`** and **`tidyr::pivot_longer()`** = transform data frame from long to wide data --- # Useful packages - janitor = miscellaneous cleaning - unheadr = weird nested headers - tidycells = multiple tables spaced out in Excel --- # janitor for cleaning columns and creating frequency tables ```r library(janitor) psm_gp %>% select(OTU, Sample, SampleType, Description) %>% filter(SampleType == "Feces") %>% mutate(Description = as.character(Description)) %>% separate_rows(Description, sep = ", ") %>% * clean_names() %>% head(5) ``` ``` ## # A tibble: 5 x 4 ## otu sample sample_type description ## <chr> <chr> <fct> <chr> ## 1 331820 M11Fcsw Feces "M1" ## 2 331820 M11Fcsw Feces "Day 1" ## 3 331820 M11Fcsw Feces "fecal swab" ## 4 331820 M11Fcsw Feces "whole body study " ## 5 331820 M31Fcsw Feces "M3" ``` --- # janitor for creating frequency tables ```r psm_gp %>% select(OTU, Sample, SampleType, Description) %>% * tabyl(SampleType) %>% * adorn_totals() ``` ``` ## SampleType n percent ## Feces 76864 0.15384615 ## Freshwater 38432 0.07692308 ## Freshwater (creek) 57648 0.11538462 ## Mock 57648 0.11538462 ## Ocean 57648 0.11538462 ## Sediment (estuary) 57648 0.11538462 ## Skin 57648 0.11538462 ## Soil 57648 0.11538462 ## Tongue 38432 0.07692308 ## Total 499616 1.00000000 ``` --- # unheadr for nested headers **Before** | scientific\_name | common\_name | red\_list\_status | mass\_kg | | :---------------------- | :--------------------------- | :---------------- | -------: | | Asia | NA | NA | NA | | CERCOPITHECIDAE | NA | NA | NA | | Trachypithecus obscurus | Dusky Langur | NT | 7.13 | | Presbytis sumatra | Black Sumatran Langur | EN | 6.00 | --- # unheadr for nested headers **After** | scientific\_name | common\_name | red\_list\_status | mass\_kg | family | | :---------------------- | :--------------------------- | :---------------- | -------: | :-------------- | | Trachypithecus obscurus | Dusky Langur | NT | 7.13 | CERCOPITHECIDAE | | Presbytis sumatra | Black Sumatran Langur | EN | 6.00 | CERCOPITHECIDAE | --- # tidycells for odd cell arrangements 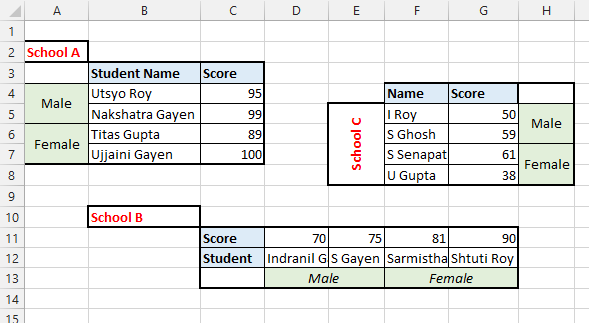 --- # Resources - R for Data Science https://r4ds.had.co.nz - Ted's list of underrated tidyverse functions https://hugo-portfolio-example.netlify.app/projects/tidyverse_functions/ - {janitor} R package https://github.com/sfirke/janitor - {unheadr} R package https://github.com/luisDVA/unheadr - {tidycells} https://r-rudra.github.io/tidycells/ - Blog post on cleaning messy data https://rfortherestofus.com/2019/12/how-to-clean-messy-data-in-r/